TAGGED: EME
-
-
June 23, 2023 at 1:17 pm
filmartinelli
SubscriberHello,
I have encountered an unexpected result in simulating the transfer matrix of a repeated unit cell with EME.Background:
I have a waveguide bragg reflector with absorbing layer and I need to calculate the absorption in each layer. To do so, I need to calculate the coefficient of the various modes in a certain position of each unit cell, but because EME does not allow to do so, I need to extract the S-matrix of the unit cell and "propagate it" in Python. In this way I can retrieve the intensity of the modes also at intermediate position.
To validate my code, I decided to propagate a simple structure with my Python code and with EME and to compare the result. The structure is a waveguide bragg reflector (no absorbing layer) consisting of two units cells. After setting up the geometry, I did the following- I calculated the S matrix of the basic unit cell with EME.
- To do so, I set the period of the unit cells to 1, I run EME propagate, and I extracted the "internal S matrix" and sent it to Python.
- I used 20 modes in the calculation
- I used the S matrix of the unit cell to calculate the S matrix of a 2-cell-structure in Python.
- The calculation of the total S matrix of two cascaded cells is an analytical solution and I verified it by comparing with litterature (here: https://ieeexplore.ieee.org/document/1125369). Thus, I am pretty sure of the correctness of the formulation.
- I calculated the total S matrix also in EME
- I used EME propagate after setting the number of periods to 2.
- I extracted the "internal S matrix"
Eventually, I compared the S matrices of the 2-unit-cells structure calculated with EME and with Python and I noticed that the results were quite different.
I have checked the correctness of the code that calculates the total S matrix and I am fairly sure of its correctness. Therefore, I had the doubt that probably I am not interpreting correctly the meaning of the "Internal S matrix" that EME propagate provides. Because the documentation about the this result and how it is obtained is limited, is there anyone that can give me an insight of what could be wrong in my reasoning?
- I calculated the S matrix of the basic unit cell with EME.
-
June 23, 2023 at 7:34 pm
Guilin Sun
Ansys EmployeeWe could not comment on Python result as it is 3rd party software.
The "internal S matrix" is the S matrix at each interface between cells, eg, the modes decomposion at both sides of the interface without any correction. Please use Lumerical script to check. It could be different operation of the matrix multiplication.
-
June 27, 2023 at 1:18 pm
filmartinelli
SubscriberI have tried to implement everything in Lumerical Script.
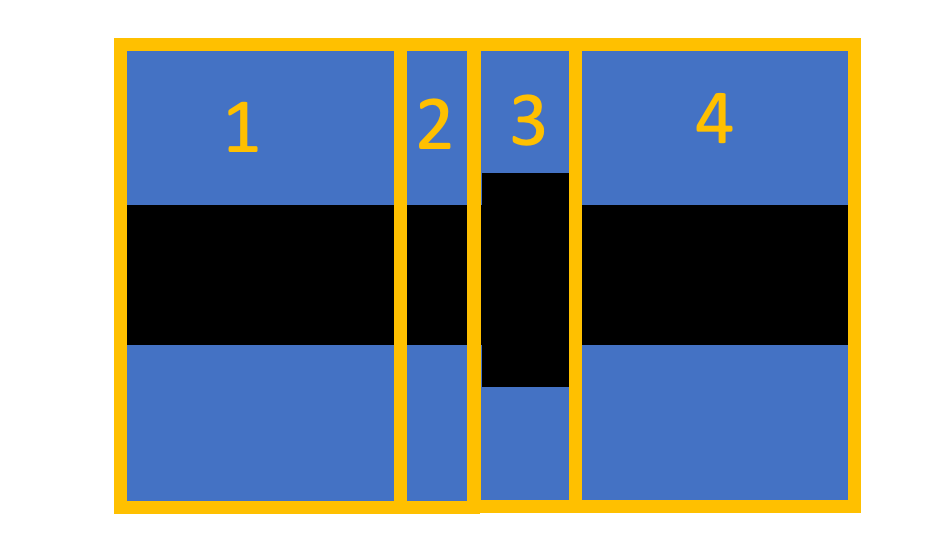
My structure looks like this:
The periodic group is set only in cells 2 and 3. To my understanding, the internal S-matrix is the S matrix from before cell 2 to after cell 3. This is based on my understanding from the Ansys video.
My Lumerical script does the following:
- Set the periods to 1 and run Eme Propagate.
- The simulated structure is [1, (2,3)^1, 4].
- Extract the Internal User Matrix. As far as I understood, this represent the S matrix of my unit cell.
- Set the periods to 2 and run EME Propagate
- The simulated structure is [1, (2,3)^2, 4].
- Extract the Internal User Matrix. At this point, this should represent the structure of two cascaded unit cells.
- Calculate the S matrix of the two cascaded unit cells analytically.
- This has been implemented with a Lumerical script function and it is a known result.
- The validity of the function has been tested by calculating the S matrix of two cascaded networks whose result was known.
- This has been implemented with a Lumerical script function and it is a known result.
Again, by comparing the S-matrix of point 2 and 3, I find quite a difference in the results.
I am pretty confident that the analytical function is correct, thus I believe there is either a mistake in the way that I interpret the S matrix or the EME algorithm is doing something beyond just cascading S matrices which I cannot find on the documentation.
I would like to have some highlights on that.Thank you
Below the script that I used:
# This function cascades two S matrices
function cascade_S(S){
rows = size(S);
rows = rows(1);
#Extract blocks
S11 = S(1:rows/2, 1:rows/2);S12 = S(1:rows/2, rows/2+1:rows);
S21 = S(rows/2+1:rows, 1:rows/2);
S22 = S(rows/2+1:rows, rows/2+1:rows);
# Identity
I = matrix(rows/2, rows/2);
for (i = 1:rows/2){I(i,i) = 1;}
# Cascade step
S11f = S11 + mult(S12, mult(S11, mult(inv(I-mult(S22, S11)), S21)));
S12f = mult(S12 , mult(inv(I-mult(S11,S22)), S12));
S21f = mult(S21 , mult(inv(I-mult(S22, S11)), S21));
S22f = S22 + mult(S21 , mult(S22, mult(inv(I-mult(S11,S22)), S12)));
# Final S
cascade = matrix(rows, rows);
cascade(1:rows/2, 1:rows/2) = S11f;cascade(1:rows/2, rows/2+1:rows) = S12f;
cascade(rows/2+1:rows, 1:rows/2) = S21f;
cascade(rows/2+1:rows, rows/2+1:rows) = S22f;
return cascade;
}### Just to test that cascade works
#x = [ -0.0011 - 0.0605i, -0.2268 + 0.1546i, -0.0967 - 0.0686i, 0.0477 + 0.0444i;
#-0.2265 + 0.1545i, -0.0693 - 0.0377i, 0.0419 + 0.0414i, -0.0998 - 0.0465i;
#-0.0961 - 0.0692i, 0.0420 + 0.0415i, -0.0499 - 0.0681i, -0.2246 + 0.1676i;
#0.0478 + 0.0445i, -0.0996 - 0.0468i, -0.2244 + 0.1673i, -0.0546 - 0.0231i];
#known_casca_x = [0.0030 - 0.0585i , -0.2312 + 0.1531i , 0.0060 + 0.0165i , -0.0053 - 0.0136i;
#-0.2309 + 0.1530i, -0.0664 - 0.0368i, -0.0042 - 0.0124i , 0.0090 + 0.0124i;
#0.0058 + 0.0166i, -0.0041 - 0.0124i, -0.0463 - 0.0669i, -0.2291 + 0.1665i;
#-0.0052 - 0.0137i, 0.0089 + 0.0124i, -0.2290 + 0.1662i, -0.0511 - 0.0223i];
#print(x);
#Sf=cascade_S(x);
#print(Sf - known_casca_x);
#print(max(abs(Sf - known_casca_x)));### ----------------- Actual code ------------------
#--------------- Point 1
setemeanalysis("override periodicity", 1);
setemeanalysis("periods", [1]);
emepropagate();
S_user_1cell = getresult("EME", "user s matrix");
S_internal_1cell = getresult("EME", "internal s matrix");rows = size(S_internal_1cell);
rows = rows(1);
idx = linspace(0,rows, rows);# Plot internal S
#image(idx, idx, abs(S_internal_1cell));# --------------- Point 2
# Set number of Cell on 2
setemeanalysis("periods", [2]);
emepropagate();
S_user_2cell = getresult("EME", "user s matrix");
S_internal_2cell = getresult("EME", "internal s matrix");# Plot internal S
#image(idx, idx, abs(S_internal_2cell));# ---------------- Point 3
# Cascade the single unit S and plot
S_cascade = cascade_S(S_internal_1cell);
#image(idx, idx, abs(S_cascade));# Plot difference between matrix in point 2 and point 3
image(idx, idx, abs(S_cascade - S_internal_2cell)); - Set the periods to 1 and run Eme Propagate.
-
June 27, 2023 at 5:03 pm
Guilin Sun
Ansys EmployeeFirst,using rows/2 can be dangeous in general, as when "rows" is an odd number, it can give you result unexpectd.
Second, you stressed that you are using "internal S matrix". however such matrix does NOT count the periodicity as it has no such information internally. It is only when emepropagate works then the periodicity is counted. Again, the "internal S matrix" only counts the interface s matrix.
To realize your goal, you can duplicate the structure to have [1, (2,3)^2, 4] after [1, (2,3)^1, 4]. Soon the reuse of cells will be available for vote so please check the ix website to vote once it is ready:
New Feature vote: Vote new features, and file your feature request
-
June 28, 2023 at 1:49 pm
filmartinelli
SubscriberHello, thank you for your reply.
I understand that probably using the "internal S matrix" is not the right choice. I have therefore reset my code such that I use the "User S Matrix".The simulation region now is the following
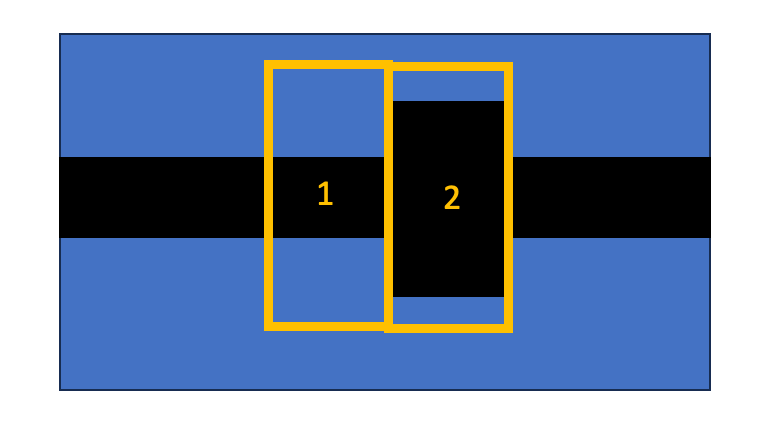
I have repeated the process that I described in the previous step.
- first I run EME Propagate for [(1,2)^1] and extract the User Matrix
- second I run EME Propagate for [(1,2)^2] and extract User Matrix. As far as I understood, the User matrix should account for the periodicity
- third I take the User Matrix of point 1 and calculate the S matrix of two cascaded systems. By right, this should be the same result as point 2. However, the two results differ again
I am not sure I fully understood the suggestion in the previous post. My goal is to get the S matrix of the single unit from EME such that I can use it in Python and cascade it N-times to access the modes field intensities in the middle of the structure. However, it seems that just cascading the S matrices extracted from EME does not give a result consistent with EME one
-
June 28, 2023 at 3:30 pm
Guilin Sun
Ansys Employee"internal s matrix" is for diagnose purpose;
"user s matrix" is a subset of the total S matrix. So when you only use it to cascade, it loses some other information. I would suggest you to find EME papers to get more details.
In your case, both will not lead to the expected result you desire. Only when you have those groups inside EME simulation can you get correct result.
-
June 29, 2023 at 9:09 am
filmartinelli
SubscriberThank you for your reply.
I understand that User S Matrix is a subset. I have included all the modes that I use in the EME in the user S Matrix such that it represent the total S matrix. i.e. I am simulating the cells with 10 modes and my user S matrix has 20 modes. If this is again not enough, I would appreciate if you could indicate me how to access the total S matrix of the system, since from Lumerial documentation I cannot understand it. -
June 29, 2023 at 5:04 pm
Guilin Sun
Ansys EmployeeUnfortunately it is not designed to give access for the total s matrix, as it has some corrections to get the "user s matrix". As I suggested, you can have the two sections cascaded. It will not take longer time than you simulate them individually, and memory might not be an issue giving the number of modes you set. This is a viable way to simulate it, and easy for you to change the number of periods.
-
- The topic ‘Unexpected S matrix from EME propagation’ is closed to new replies.



-
3432
-
1057
-
1051
-
902
-
896

© 2025 Copyright ANSYS, Inc. All rights reserved.