TAGGED: ls_dyna
-
-
July 27, 2023 at 9:21 pm
mehdisaloo
SubscriberHello,
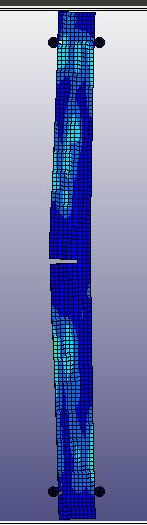
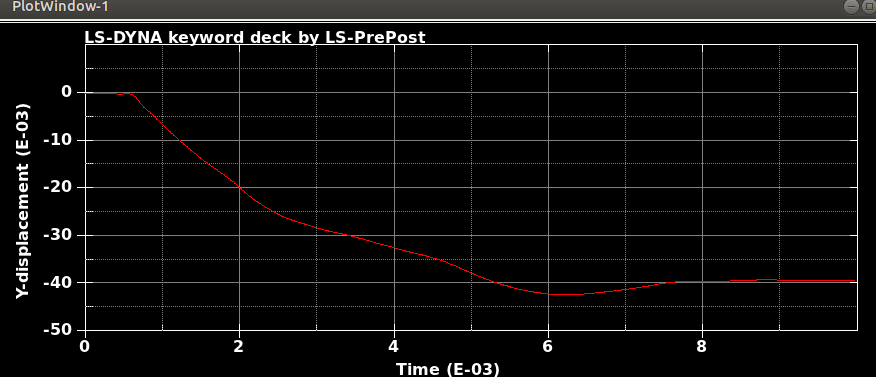
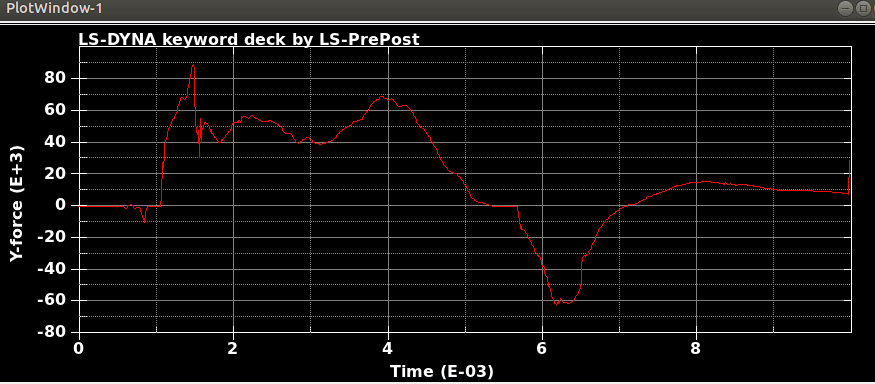
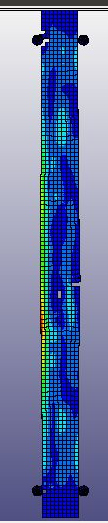
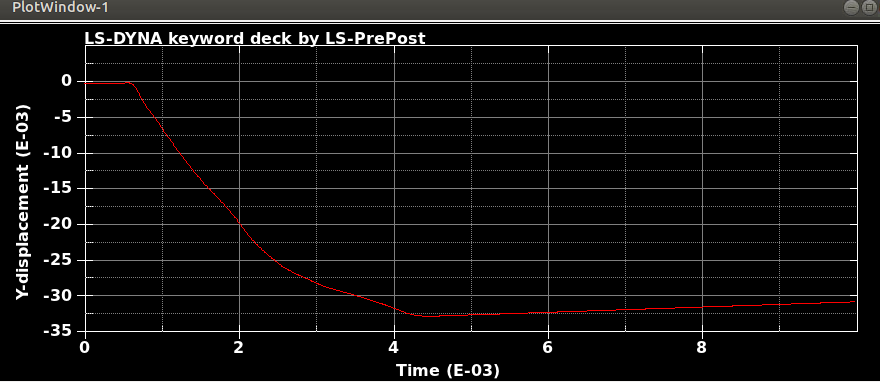
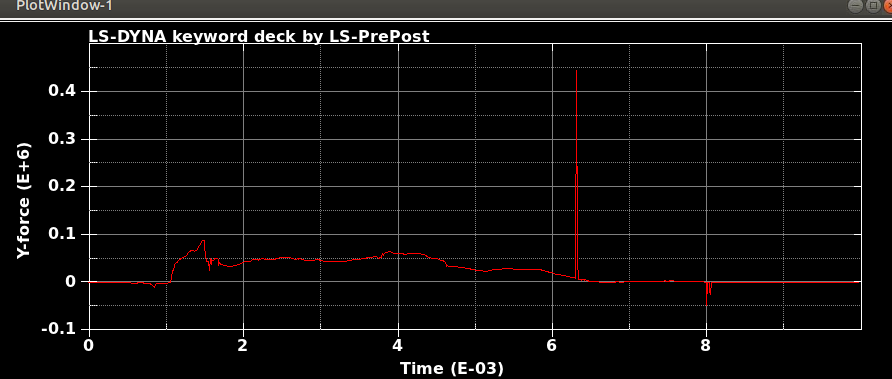
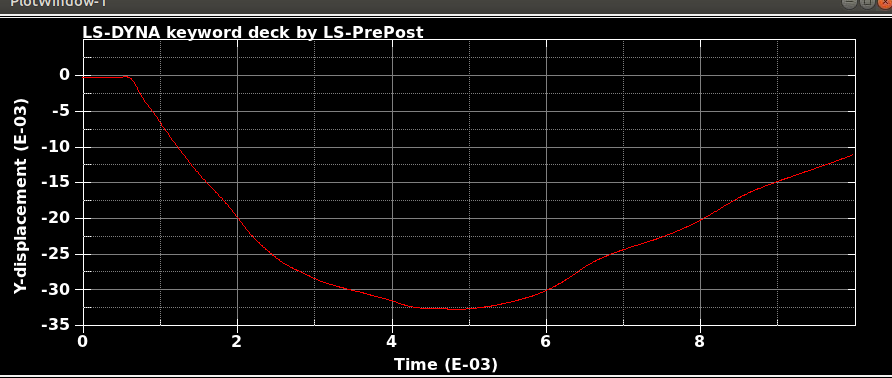
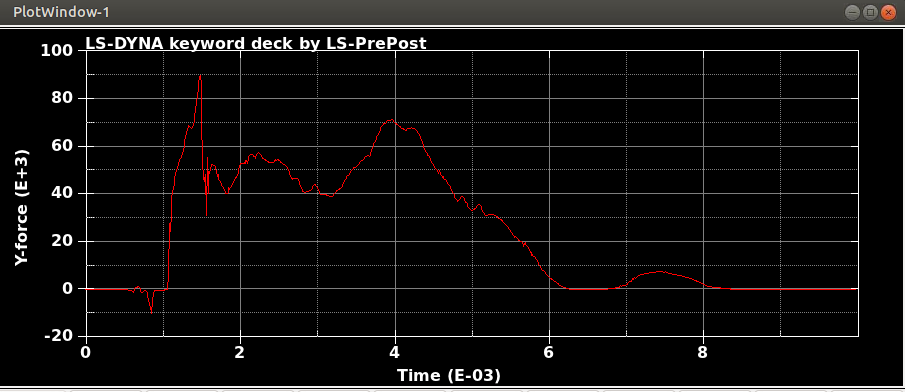
When I run the same file in the same version of LS-DYNA, I will get different results! I mean, I just copy and run the model in another directory. So, I expect to see the same results. It is LS-DYNA LS-Run 2023 R1 (smp s R13) and I use the same NCPU and MEMORY. I get different results even if I run the model in the same directory again. In that case, the software will delete all results files and write the new results. I emphasize that the model is exactly the same. You can see the different results, especially for damage patterns from the same model as follows.
I would appreciate it if you could give me some advice.
Thanks in advance.
Regards,
Mehdi
Results #1
Results #2
Results #3
-
July 28, 2023 at 4:01 pm
Reno Genest
Ansys Employee-
July 31, 2023 at 7:36 pm
mehdisaloo
SubscriberHello Reno,
Thanks ever so much for your help, I really appreciate it.
I used consistency checking with 4 cores by using ncpu=-4 and SMP single-precision solver. In that case, repeated runs then produce the same results.
I also tried with a double precision solver and ncpu=-4. In that case, repeated runs produce the same results as well, although it is significantly different from the SMP single-precision solver.
Therefore, both single and double precision solvers give me consistent results but there is a significant difference between them. I wonder what the benefit of a double precision solver is and if it leads to better and more accurate results.
I would appreciate it if you could give me some advice.
Thanks in advance.
Regards,
Mehdi
-
-
July 31, 2023 at 7:43 pm
Reno Genest
Ansys EmployeeHello Mehdi,
If the results from single precision are different than with double precision, then it means you need to run double precision. I always run double precision. Here is more information from our knowledge database:
”I almost always use double precision. The only way to know with any degree of certainty whether single precision is adequate or not is to compare the single precision solution to the double precision solution. If there is good agreement, then single precision may be deemed adequate for that particular model (and likely so for other models that are similar).”
Let me know how it goes.
Reno.
-
July 31, 2023 at 11:17 pm
mehdisaloo
SubscriberHello Reno,
Thanks so much for your swift response.
I need, therefore, always set ncpu= to a negative number of cores (e.g. ncpu=-4) and run double precision in order to get consistent results.
I wanted to ask you one more technical question regarding maintaining consistency. I have read LS-Run User's Guide and found the following relevant information.
“When you are using shared memory parallel processing, the calculations may be executed in different order, depending on CPU availability and the workload on each CPU. Because of this, you may see slight differences in the results when you run the same job multiple times. To avoid these differences, you can specify the number of CPUs as a negative number to maintain consistency. Maintaining consistency can result in an increase of up to 15% in CPU time.”
As far as I understand, when I specify to use multiple cores (e.g. ncpu=4), I am running using parallel processing (i.e. multiple cores), thus getting inconsistent results is probable. I set the number of cores to 1 in LS-DYNA (ncpu=1) to see if repeated runs then produce the same results. In that case, I get inconsistent results, however, I am not running using parallel processing! (when I set ncpu=-1, I get consistent results.)
I would appreciate it if you could give me some advice.
Thanks in advance.
Regards,
Mehdi
-
-
August 1, 2023 at 3:51 pm
Reno Genest
Ansys EmployeeHello Mehdi,
When the consistency flag is not used (ncpu= positive number or by setting CONST=2 on *CONTROL_PARALLEL), summation of quantities (results) occur in a different order between runs. This leads to some randomness in the round-off error. For a model with failure and element deletion as in your case, this randomness can lead to bifurcation in the solution. If in a given run, the failure strain is reached sooner for a given element, then this element will get deleted sooner, which in turns will increase the strain on the other remaining elements near the element that was deleted. For this reason, a given run may lead to more elements being deleted compared with another run, even though you are running on 1 CPU.
When the consistency flag is activated (ncpu= negative number or CONST=1 on *CONTROL_PARALLEL), the summation of quantities is done in a precise order so that the round-off error is the same between runs. This ensure nearly the same results between runs.
You will find more information about consistency in the LS-DYNA User Manual Vol I in remark 2 of the *CONTROL_PARALLEL keyword:
You can download the LS-DYNA user manuals here:
https://lsdyna.ansys.com/manuals/
Reno.
-
August 1, 2023 at 5:10 pm
mehdisaloo
SubscriberHello Reno,
I grasped the points you mentioned. Thanks ever so much for your help, I really appreciate it.
Regards,
Mehdi
-
-
- The topic ‘Same File Different Results!’ is closed to new replies.



-
4708
-
1565
-
1386
-
1242
-
1021

© 2026 Copyright ANSYS, Inc. All rights reserved.