Ansys Learning Forum › Forums › Discuss Simulation › Photonics › Unexpected S matrix from EME propagation › Reply To: Unexpected S matrix from EME propagation
I have tried to implement everything in Lumerical Script.
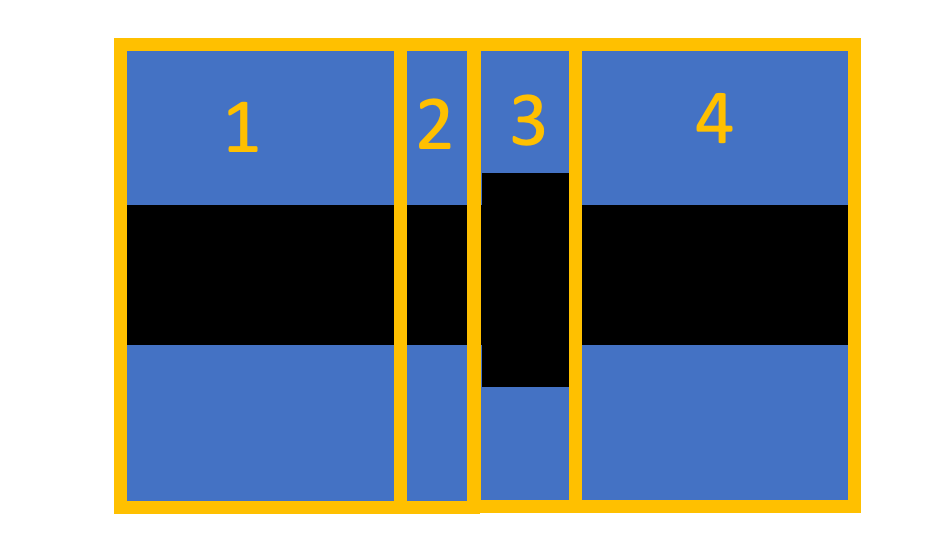
My structure looks like this:
The periodic group is set only in cells 2 and 3. To my understanding, the internal S-matrix is the S matrix from before cell 2 to after cell 3. This is based on my understanding from the Ansys video.
My Lumerical script does the following:
- Set the periods to 1 and run Eme Propagate.
- The simulated structure is [1, (2,3)^1, 4].
- Extract the Internal User Matrix. As far as I understood, this represent the S matrix of my unit cell.
- Set the periods to 2 and run EME Propagate
- The simulated structure is [1, (2,3)^2, 4].
- Extract the Internal User Matrix. At this point, this should represent the structure of two cascaded unit cells.
- Calculate the S matrix of the two cascaded unit cells analytically.
- This has been implemented with a Lumerical script function and it is a known result.
- The validity of the function has been tested by calculating the S matrix of two cascaded networks whose result was known.
- This has been implemented with a Lumerical script function and it is a known result.
Again, by comparing the S-matrix of point 2 and 3, I find quite a difference in the results.
I am pretty confident that the analytical function is correct, thus I believe there is either a mistake in the way that I interpret the S matrix or the EME algorithm is doing something beyond just cascading S matrices which I cannot find on the documentation.
I would like to have some highlights on that.
Thank you
Below the script that I used:
# This function cascades two S matrices
function cascade_S(S){
rows = size(S);
rows = rows(1);
#Extract blocks
S11 = S(1:rows/2, 1:rows/2);
S12 = S(1:rows/2, rows/2+1:rows);
S21 = S(rows/2+1:rows, 1:rows/2);
S22 = S(rows/2+1:rows, rows/2+1:rows);
# Identity
I = matrix(rows/2, rows/2);
for (i = 1:rows/2){I(i,i) = 1;}
# Cascade step
S11f = S11 + mult(S12, mult(S11, mult(inv(I-mult(S22, S11)), S21)));
S12f = mult(S12 , mult(inv(I-mult(S11,S22)), S12));
S21f = mult(S21 , mult(inv(I-mult(S22, S11)), S21));
S22f = S22 + mult(S21 , mult(S22, mult(inv(I-mult(S11,S22)), S12)));
# Final S
cascade = matrix(rows, rows);
cascade(1:rows/2, 1:rows/2) = S11f;
cascade(1:rows/2, rows/2+1:rows) = S12f;
cascade(rows/2+1:rows, 1:rows/2) = S21f;
cascade(rows/2+1:rows, rows/2+1:rows) = S22f;
return cascade;
}
### Just to test that cascade works
#x = [ -0.0011 - 0.0605i, -0.2268 + 0.1546i, -0.0967 - 0.0686i, 0.0477 + 0.0444i;
#-0.2265 + 0.1545i, -0.0693 - 0.0377i, 0.0419 + 0.0414i, -0.0998 - 0.0465i;
#-0.0961 - 0.0692i, 0.0420 + 0.0415i, -0.0499 - 0.0681i, -0.2246 + 0.1676i;
#0.0478 + 0.0445i, -0.0996 - 0.0468i, -0.2244 + 0.1673i, -0.0546 - 0.0231i];
#known_casca_x = [0.0030 - 0.0585i , -0.2312 + 0.1531i , 0.0060 + 0.0165i , -0.0053 - 0.0136i;
#-0.2309 + 0.1530i, -0.0664 - 0.0368i, -0.0042 - 0.0124i , 0.0090 + 0.0124i;
#0.0058 + 0.0166i, -0.0041 - 0.0124i, -0.0463 - 0.0669i, -0.2291 + 0.1665i;
#-0.0052 - 0.0137i, 0.0089 + 0.0124i, -0.2290 + 0.1662i, -0.0511 - 0.0223i];
#print(x);
#Sf=cascade_S(x);
#print(Sf - known_casca_x);
#print(max(abs(Sf - known_casca_x)));
### ----------------- Actual code ------------------
#--------------- Point 1
setemeanalysis("override periodicity", 1);
setemeanalysis("periods", [1]);
emepropagate();
S_user_1cell = getresult("EME", "user s matrix");
S_internal_1cell = getresult("EME", "internal s matrix");
rows = size(S_internal_1cell);
rows = rows(1);
idx = linspace(0,rows, rows);
# Plot internal S
#image(idx, idx, abs(S_internal_1cell));
# --------------- Point 2
# Set number of Cell on 2
setemeanalysis("periods", [2]);
emepropagate();
S_user_2cell = getresult("EME", "user s matrix");
S_internal_2cell = getresult("EME", "internal s matrix");
# Plot internal S
#image(idx, idx, abs(S_internal_2cell));
# ---------------- Point 3
# Cascade the single unit S and plot
S_cascade = cascade_S(S_internal_1cell);
#image(idx, idx, abs(S_cascade));
# Plot difference between matrix in point 2 and point 3
image(idx, idx, abs(S_cascade - S_internal_2cell));