-
-
February 14, 2022 at 4:47 am
jordankim
SubscriberI'm trying to add a new structure to Lumerical using the command 'addplanarsolid'.
In detail, following the 'Drawing truncated cone in finite element.lsf', I have understood the script and converted to Python in order to conduct it by Python.
I understood that the command 'addplanarsolid' requests two data i.e. vertices and facets.
vtx = [[ 3.00000000e-06 1.50000000e-06 -1.50000000e-06 -3.00000000e-06
-1.50000000e-06 1.50000000e-06 1.50000000e-06 7.50000000e-07
-7.50000000e-07 -1.50000000e-06 -7.50000000e-07 7.50000000e-07
0.00000000e+00 0.00000000e+00]
[ 0.00000000e+00 2.59807621e-06 2.59807621e-06 3.67394040e-22
-2.59807621e-06 -2.59807621e-06 0.00000000e+00 1.29903811e-06
1.29903811e-06 1.83697020e-22 -1.29903811e-06 -1.29903811e-06
0.00000000e+00 0.00000000e+00]
[-5.00000000e-06 -5.00000000e-06 -5.00000000e-06 -5.00000000e-06
-5.00000000e-06 -5.00000000e-06 -5.00000000e-06 -5.00000000e-06
-5.00000000e-06 -5.00000000e-06 -5.00000000e-06 -5.00000000e-06
-5.00000000e-06 5.00000000e-06]]
a = [[[2, 1, 13]], [[3, 2, 13]], [[4, 3, 13]], [[5, 4, 13]], [[6, 5, 13]], [[1, 6, 13]], [[7, 8, 14]], [[8, 9, 14]], [[9, 10, 14]], [[10, 11, 14]], [[11, 12, 14]], [[12, 7, 14]], [[1, 2, 8, 7]], [[2, 3, 9, 8]], [[3, 4, 10, 9]], [[4, 5, 11, 10]], [[5, 6, 12, 11]], [[6, 1, 7, 12]]]
I have made the vertex and facets then I encountered the error
"File "E:/user/김강석/lumapi_example/pyramid.py", line 121, in
fdtd.set('facets', a)
lumapi.LumApiError: "argument list for set command is incorrect - set arguments were converted to ('str, cell array)"
So I converted arguments of facets "a" to nd.array and I also encountered the error like this
"E:/user/김강석/lumapi_example/pyramid.py:114: VisibleDeprecationWarning: Creating an ndarray from ragged nested sequences (which is a list-or-tuple of lists-or-tuples-or ndarrays with different lengths or shapes) is deprecated. If you meant to do this, you must specify 'dtype=object' when creating the ndarray
a_array = np.array(a)
TypeError: float() argument must be a string or a number, not 'list'
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "C:Program FilesLumericalv202apipythonlumapi.py", line 362, in packMatrix
v = v.astype(dtype="float64", casting="unsafe", order='F')
ValueError: setting an array element with a sequence.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "E:/user/김강석/lumapi_example/pyramid.py", line 121, in
fdtd.set('facets', a_array)
...
lumapi.LumApiError: 'Unsupported data type'
Process finished with exit code 1
If I resolved this problem, then should I put the command 'a_array = fdtd.putv("a",a)' like this?
Any help would be appreciated
February 14, 2022 at 10:47 pmKyle
Ansys Employee
I think the issue here is with the data types you're using. The facets variable "a" in this example is a cell array containing cell arrays that contain matrices. In Python this would be a list of lists of NumPy arrays, while in your first example it is a list of lists of lists. So you should change the final level from lists to NumPy arrays.
However this is very confusing. There are two methods of defining the facets, as described on the Planar solid documentation page. You can define them as cell arrays, like in this example, or as matrices. I would recommend you write the facets using matrices (or, in Python, as NumPy arrays) instead of cells. This should be much simpler, I think.
February 17, 2022 at 1:59 amjordankim
SubscriberThank you for your advice
I have tried to convert the facets variable "a" to matrix, before that I also have converted the facets variable in "Planar solid" script from cell array to matrix in order to apply this method to our project.
I'm finally do it, so I don't have no error code in my python script.
But As I open the lumerical fsp file from python file, I can't see Any completed solid structure like this image
Any help would be appreciated
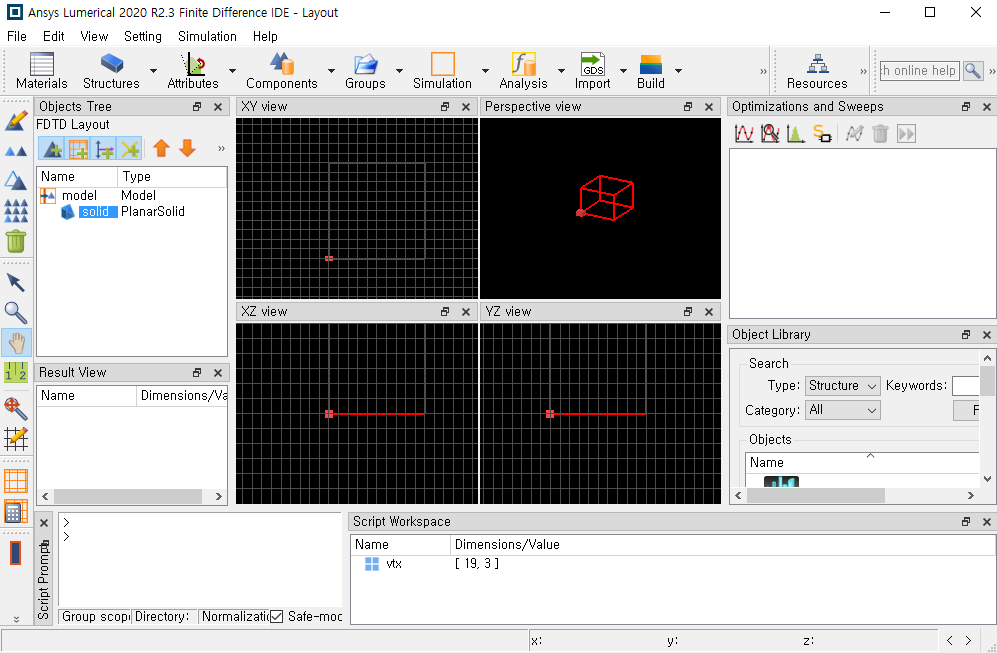
facet(matrix)
[[ 1.9. 15.]
[ 4. 10. 16.]
[ 3. 11. 17.]
[ 2.0.0.]]
[[1. 0. 0.]
[5. 0. 0.]
[8. 0. 0.]
[4. 0. 0.]]
.
.
.
[[15.0.0.]
[17.0.0.]
[20.0.0.]
[18.0.0.]]
[[17.0.0.]
[16.0.0.]
[19.0.0.]
[20.0.0.]]
As a result of checking, I think it is same in lumerical script facet matrix
February 22, 2022 at 9:56 pmKyle
Ansys Employee
A red box like this indicates that there is some error in a simulation object, usually because of an error in an analysis/structure group script. In this case it looks like this is because of an error encountered by the planar solid object. I would recommend prototyping your script in an LSF script file to make sure everything works before converting it to Python. Ultimately it might be easier to just use the LSF script to create the object and call the script from Python using the lumapi.eval method: lumapi.eval("script_name.lsf")
January 7, 2023 at 2:50 pmmartinmi
SubscriberHallo, i just try to do the same in Lumerical and get similar results as @jordankim. I try to generate the same structure with LSF file, just to verify if my code (structure) is correct. Witht the MatLab LSF file format, everything is working and i can generate the structure. Did you resolved the issiu in some way, and if you did, can you post some python example ?
Viewing 4 reply threads- The topic ‘Adding new structure to lumerical via Python using ‘addplanarsolid’’ is closed to new replies.
Ansys Innovation SpaceTrending discussionsTop Contributors-
3862
-
1414
-
1220
-
1118
-
1015
Top Rated Tags© 2025 Copyright ANSYS, Inc. All rights reserved.
Ansys does not support the usage of unauthorized Ansys software. Please visit www.ansys.com to obtain an official distribution.
-


Ansys Assistant

Welcome to Ansys Assistant!
An AI-based virtual assistant for active Ansys Academic Customers. Please login using your university issued email address.
Hey there, you are quite inquisitive! You have hit your hourly question limit. Please retry after '10' minutes. For questions, please reach out to ansyslearn@ansys.com.
RETRY







