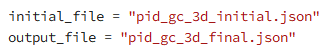-
-
December 2, 2024 at 11:15 am
rahul.sanghvi
SubscriberIn step 2 of 2D grating coupler design, how is the sweep length determined ?
I was expecting the source sweep would be carried out over the entire length of the grating, but in the example file it sweeps from 4 µm to 7 µm only.
-
December 3, 2024 at 6:00 am
anna.wirth-singh
Ansys EmployeeHi Rahul,
Thanks for your question. Could you please clarify which example you are working with? We have a few grating coupler design examples, so I want to check that I am looking at the right one to answer your question.
We have one here using the built-in particle swarm optimization tool: Grating coupler – Ansys Optics
This one uses the Python-based inverse design package lumopt: Inverse Design of Grating Coupler (2D) – Ansys Optics
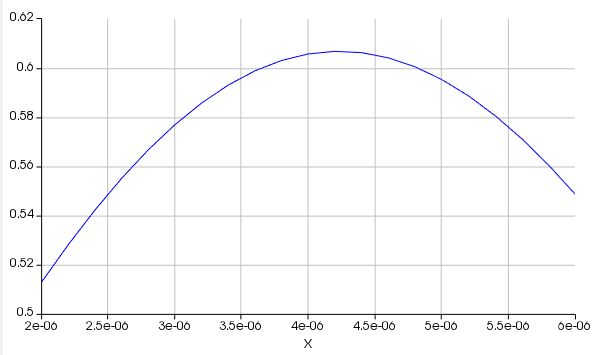
For a sweep over source position, there is typically a well-defined maximum. The sweep over source position probably looks something like this, where the y-axis is the transmission into the waveguide and the x-axis is the source position:
For this parameter, it is clear that the optimal value is somewhere near the center. Most likely, the designer of the example file restricted the sweep range so that they could run fewer points (and therefore get results faster), with the knowledge that the optimal value would not be near the endpoints of the sweep.
-
December 3, 2024 at 9:37 am
rahul.sanghvi
SubscriberHi Anna,
I am refering to the inverse design example: https://optics.ansys.com/hc/en-us/articles/360042800573-Inverse-Design-of-Grating-Coupler-2D
and the code "pid_grating_coupler_sweep_2D.py" in the second step.
I am not able to follow how did the designer define the start and the end point of the sweep. I would like to determine the transmission over the entire length of the grating.
-
December 3, 2024 at 10:15 am
rahul.sanghvi
SubscriberHi Anna,
I am wondering if you could help me out with implementing optimisation based FDTD simulations on university supercomputer.
Currently, I am using a desktop where I open the lumerical GUI and load a python file in the “script file editor” section and execute it.
To implement this on the supercomputer, we do not have a visual node where I can do the same process.
Can you suggest a method where I run this python file in the supercomputer terminal and it somehow does the FDTD simulation -
December 3, 2024 at 11:06 pm
anna.wirth-singh
Ansys EmployeeHi Rahul,
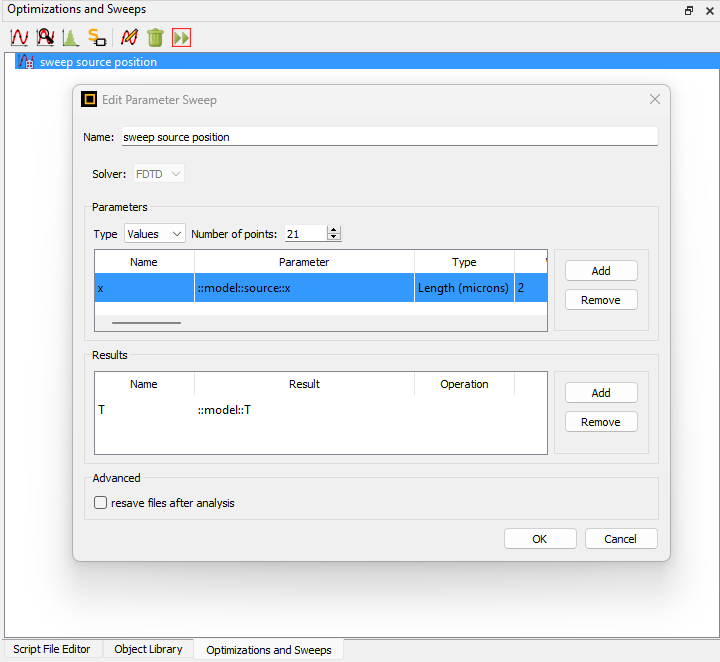
Thanks for the additional information. For your first question regarding the sweep, the script “pid_grating_coupler_sweep_2D.py” is calling a sweep that is defined in the base Lumerical file "pid_grating_coupler_2D_TE_base .fsp". Specifically, if you look at line 85 in the Python script, it uses runsweep - Script command – Ansys Optics to run the sweep that has previously been defined in the base Lumerical file. So the sweep endpoints are set in the base Lumerical file. On the ‘Optimizations and Sweeps’ tab, you should see the sweep ‘sweep source position.’ From there, you can edit the values that are included in the sweep. This is illustrated in the screenshot below:
Regarding your second question about running FDTD on a high-performance computer, I can direct you to some resources to get started. Please see this page for a general overview: Introduction to High-Performance Computing with Lumerical – Ansys Optics
It sounds like you will be running on a local cluster, so this page Running Lumerical on a Local On-premise Cluster – Ansys Optics contains instructions in how to set this up.
Best,
Anna
-
December 5, 2024 at 1:35 pm
rahul.sanghvi
SubscriberHi Anna,
Thanks for explaining. The first answer regarding sweep is clear.
Regarding the cluster, the given example describes about a sweep where the .fsp files are created, saved and then FDTD is performed. How can I run the file "pid_grating_coupler_2D_1etch.py" in the same way ?
It does not create any .fsp file but performs FDTD upto 100 iterations (or any user defined number) to optimise the grating structure. In this case, how can I perform this optimsation on HPC.
-
December 10, 2024 at 1:11 pm
rahul.sanghvi
SubscriberHi Anna,
Looking forward to hearing from you.
Thanks in advance
-
December 10, 2024 at 11:20 pm
Lito
Ansys Employee@rahul.sanghvi,
To run Python API will require a GUI connection and a GUI license. >>Python API overview – Ansys Optics
If you do not have a GUI connection (headless Linux system);
- Consult with your IT/cluster admins to:
- Install or configure the MPI - used to run FDTD simulations (.fsp) with Lumerical 2024 releases >>Running simulations with MPI on Linux – Ansys Optics
- Configure your environment for Lumerical API >>How to run Lumerical API scripts from terminal in Linux – Ansys Optics
- And run with virtual display >>Running CAD jobs on headless Linux systems – Ansys Optics<<
- Where on the newer versions you simply set QT_QPA_PLATFORM=OFFSCREEN and run the API (.py) script.
Hope this helps.
Lito
-
December 11, 2024 at 8:51 am
rahul.sanghvi
SubscriberHi Lito,
The university cluster has both, GUI and engine licence.
Earlier, I have tried out the links that you have mentioned but I could not make it work. I dont have much programming knowledge thats why I am struggling with the set up.
Nevertheless, I will try again.
Thanks
-
December 11, 2024 at 11:04 am
-
December 11, 2024 at 9:04 pm
Lito
Ansys Employee@rahul.sanghvi,
If you set the environment for Lumerical API in the cluster then you simply remove the PATH set in your script.
Set environment to Lumerical 2024 R2.3 using the default install path and our bundled Python3: (change the “install path“ according to the path in your cluster)
export PATH=/opt/lumerical/v242/bin:/opt/lumerical/v242/python/bin:$PATH
export PYTHONPATH=$PYTHONPATH:/opt/lumerical/v242/api/python
export QT_QPA_PLATFORM=offscreenExample Python Script: save as lumtest.py, – without the path settings.
import lumapi
fdtd = lumapi.FDTD(hide=True)
fdtd.addfdtd()
fdtd.addring()
fdtd.addmesh()
fdtd.addrect()
fdtd.save("mytest.fsp")Then run the example script above in the cluster:
cd /{directory/folder of you script}
python lumtest.pyIf it creates (mytest.fsp) on the same folder as your script, then this confirms that you can run Lumerical API on the cluster.
-
January 9, 2025 at 12:06 am
rahul.sanghvi
SubscriberIn 3D grating coupler design (refering to the link below), the codes pid_gc_3d_sweeps.py and pid_gc_3d_optimization.py create a new .fsp file for simulations. Can you suggest a method where I use these optimisation algorithms for an existing .fsp file.
I have my 3D grating coupler already designed but I want to optimise it further.
Thanks in advance.
(https://optics.ansys.com/hc/en-us/articles/1500000306621-Inverse-design-of-grating-coupler-3D) -
January 9, 2025 at 12:46 am
anna.wirth-singh
Ansys EmployeeHi Rahul,
Thanks for your question. In this example, the initial design is provided by parameters in a .json file (see these lines in pid_gc_3d_optimization.py) :
One option is to create a .json file corresponding to your desired initial parameters, and specify that file as the initial file here.
Alternatively, some of our other lumopt examples load the initial conditions from an existing .fsp or setup script. For example, in Inverse design of waveguide crossing – Ansys Optics, crossing_opt_3D.py loads the initial simulation from lsf script:
I think that you could similarly load a base simulation for the grating coupler. I will note, however, that the optimizable geometry typically needs to be defined separately as a FunctionDefinedPolygon with parameters that are optimized. So the base simulation setup is only for the input/output waveguides, mesh override region, and FOM monitors, while the optimizable geometry is constructed from a Python script using supplied initial parameters. So in either case, the optimizable geometry needs to be defined by parameters (numbers) that are optimized.
I hope this helps.
Best,
Anna
-
January 14, 2025 at 1:18 pm
rahul.sanghvi
SubscriberHi Anna,
In 2D inverse design grating coupler, the grating position are defined by 50 points/cordinates. This result is used in 3D inverse design grating coupler but it has now 51 points/cordinates. Can you explain how is there one additional cordinate ?
Please refer to files:
2D inverse design grating coupler: pid_optim_final.json
3D inverse design grating coupler: pid_gc_2d_initial.json -
January 21, 2025 at 2:48 pm
rahul.sanghvi
SubscriberHello,
I modified the Inverse design of grating coupler (2D) according to my purpose, everything worked as expected. But in the last step "pid_grating_coupler_2D_1etch.py" my optimisation gets stuck after the first simulation.
I tried changing the bounds, pgtol, and the step size, but it still does not work.
Can you point out what could be wrong here ?
Thanks in advance!
-
January 29, 2025 at 11:04 pm
anna.wirth-singh
Ansys EmployeeHi Rahul,
Apologies for the delayed reply - I was on business travel.
Often, the reason that an optimization gets stuck is the optimizable geometry wanders outside the simulation region or outside the mesh override. Can you please check that the mesh override is covering the entire structure?
Best,
Anna
-
February 26, 2025 at 12:34 pm
-
February 27, 2025 at 1:21 pm
rahul.sanghvi
SubscriberHi Anna,
Can you tell me how to run LMS codes on HPC ?
For FDTD I use fdtd-engine-impi-lcl
What is the equivalent for LMS files ?Thanks in advance.
-
February 27, 2025 at 3:32 pm
Lito
Ansys EmployeeHi Rahul,
To run MODE (.lms) files from the command line/terminal use the equivalent solver executable/binary. (see this KB)
fdtd-engine-impi-lcl
varfdtd-engine-impi-lcl
eme-engine-impi-lcl
-
March 3, 2025 at 9:22 am
rahul.sanghvi
SubscriberThank you for the information. Can you guide me about the issue with gds file conversion.
-
March 3, 2025 at 2:41 pm
rahul.sanghvi
SubscriberI downloaded the 2D inverse design grating coupler from the link attahced below.
link: https://optics.ansys.com/hc/en-us/articles/360042800573-Inverse-Design-of-Grating-Coupler-2DWhen I tried to open the 3D GDS file "pid_grating_coupler_3d", I got the following error (image attahced). How to resolve this ?

-
March 6, 2025 at 2:12 pm
Kirill
Forum ModeratorDear Rahul,
Please check the topic Exporting Inverse Design of a Grating to GDS for relevant information.
Also, kindly keep discussions on the same topic within a single thread. If you have a new question, please start a new thread and reference previous discussions if relevant.Best regards,
Kirill
-
- You must be logged in to reply to this topic.



-
5094
-
1831
-
1387
-
1248
-
1021

© 2026 Copyright ANSYS, Inc. All rights reserved.